3 Methods for Modeling Protein Aggregation
How do clumps of bad proteins form? A few different ways...
Continuing on my bioinformatics kick, I recently read a fascinating NIH paper about about polymerization, which we can think of as cellular construction. Protein chains, which I have written about previously, serve two vital purposes in our cells. First, they create a cellular skeleton that gives the cell its shape. Second, they form networks of roads and highways that allow various cellular components to move around.
The cell carefully controls how these protein highways and supports are built using special helper molecules. Some of these helpers create branches in the protein chains. Others act like anchors, connecting these protein networks to the cell's outer boundary. But sometimes things can go wrong, proteins can start clumping together inappropriately. One of the most well-known examples of this happens in Alzheimer's disease, where proteins called beta-amyloid create harmful clumps in the brain that block normal cellular function.
Scientists have developed different ways to understand how these protein clumps form. Which I'll investigate here:
In the first and most basic mode, individual proteins join an existing clump, eventually reaching a physical limit.

Under this model, we can see that the aggregate reaches equilibrium before the proteins are exhausted. This points to the slow rate at which the aggregates grow and that they cannot grow infinitely through time even at large concentrations of monomers. But it doesn't explain the creation of small aggregates.
To that end, the second scenario, or the autocatalytic model was developed. In this model the aggregation happens in a chain reaction in which changed proteins can only join with other changed proteins to form clumps.

The Kinetics of the model is similar to the previous one, exhaustion of monomers happens at a fast rate but the amount of proteins capable to create aggregates also rises to the same level.
So here we have a third scenario, which involves a middle step. Before proteins can join the clump, they need to go through a change, this actually slows down the clumping process because there's a limit to how quickly proteins can go through this transformation.
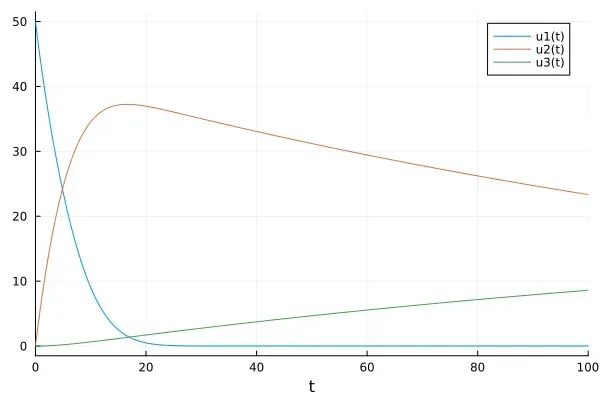
Understanding these different ways that proteins can assemble or clump together helps scientists develop treatments for diseases caused by protein clumping. It also helps us appreciate how cells normally keep these processes under control to build useful structures rather than harmful clumps.
The key difference between normal protein assembly (polymerization) and harmful clumping (aggregation) is organization. When proteins assemble normally, everything has its proper place. When proteins clump abnormally, there is chaos, though scientists are often discovering hidden patterns in the chaos.