Impact of Genetic Mimicry on COVID-19 Adaptability
The main idea behind
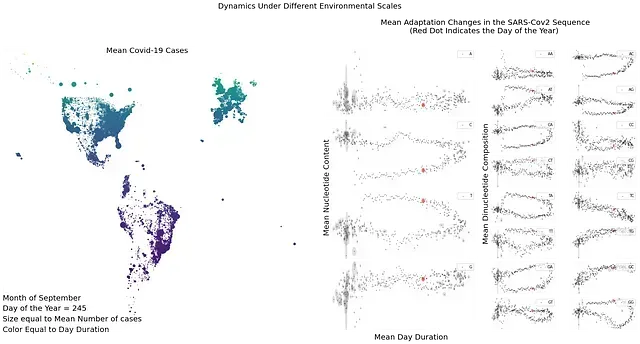
Cyclical components inside SARS Cov 2 sequence hint at a deterministic pattern inside the sequence. This characteristic must be driven by some deterministic factors. Those factors could be the environment, host, or host adaptations to the environment. By following some of those variables SARS Cov-2 is able to adapt and continuously infect the host. Through this adaptation process, small changes in the sequence will occur leading to new variants. Although this process may appear random, this could be an effect that we are unaware of as the way viruses adapt to the host.

If we look at the SARS Cov2 as a foreign invader trying to colonize a new location. Its capability to colonize the new land will depend on the available natural resources. As the virus is a parasite that relies on the molecular machinery of the host to make copies of itself, then there are two main resources needed. The nucleotides, that are needed to make copies of the genetic material. And the amino acids that are used to synthesize the different proteins needed for its assembly.
This dependence on the resources could explain the cyclical pattern inside the sequence. If we assume that host adaptations to the environment as the main driving force for viral adaptation. Then the virus will try to mimic the gene pool or transcript pool composition of the host at any given time. Or the host will be more susceptible to viral infection at times when the nucleotide pool matches the genetic composition of the virus.
Previous scenarios offer some level of explanation to a previously described phenomenon, endemicity. Tightly following the nucleotide pool can lead to a sustained level of susceptibility. Reaching a plateau after some time, a hypoendemic scenario. While the burst of high susceptibility will fall in line with periods of time with high similarity between the virus and the nucleotide pool, a hyperendemic scenario.
Recapitulating, the environment change, this change generates a response in the host. Leading to the turning on and off of some seasonal genes. Changes in gene expression patterns will in turn change the availability of the resources needed for the synthesis of viral particles. These changes will make a cell susceptible to an infection at particular environmental conditions are met.
A possible mechanism
When the SARS Cov2 virus reaches the cell it internalizes to the cell. Once inside replication of the viral genetic material leads to generating a -RNA sequence. This negative sequence is used as a template to later synthesize +RNA copies. Another process that takes place during the infection process is the shut down of gene expression. This can be achieved by the degradation of cytoplasmic mRNA. In the case of SARS Cov2, the nuclease Nsp15 crops the different host mRNA into pieces. There is a lot more going on during the infection but mi just going to focus on these two processes.
On the side of the host, the first action against the infection is the degradation of nucleotides. This action will lower the free nucleotides, aiming to starve the virus. Without nucleotides, the virus is unable to replicate its genetic material. Unless it uses an alternative source. If free nucleotides are not available or become scarce. Then the only remaining source of nucleotides will be the mRNA fragments.
Capturing the fragments could follow a mechanism like the annealing of primers in a PCR reaction. Local changes in temperature linearize the viral -RNA template, then the different fragments hybridize with the template. Or at the same time as the -RNA template is being synthesized, the available fragments start to hybridize with the -RNA. This will lead to a series of spots that later are filled with the available nucleotides.
This possible replication mechanism would be able to explain the cyclical patterns. And offer an explanation for the seasonality of viral diseases. It could also explain why the use of fragment frequency to analyze the sequences can capture such behavior. Recombination and mutation could be an intermediate step between two environmental conditions. As the available fragment pools changes, then different kinds of annealings and gaps are possible.
As a whole, the sequence contains the same components. But different arrangements lead to new changes in amino acids. Resulting in the new variants, immune response, and evasion, and not because the virus is aiming to do it. But because those will be the only viral constructs that will survive long enough to infect another host.
This mechanism could also explain the development of autoimmunity due to viral infection. When a large enough fragment is added to a structural element of the virus. Then such fragment will be recognized as foreign. Later when the infection is cleared, the immune system will be able to recognize such fragments and react to them.
Some evidence of mimicry
Reconstructing the SARS Cov2 sequence from a series of fragments is a complicated task. On one side is the fact that as the SARS Cov2 evolves, generating changes in its sequence. This in turn also changes the number and kind of similar fragments. Thus a single sequence comparison will lead to a biased approximation. Another problem is the lack of seasonal gene expression databases. (or at least not to my knowledge) This increases the number of comparisons. Thus on the host side of things the best option is to compare the SARS Cov2 sequences with the reference transcripts. This will approximate the available fragment and nucleotide pool inside the cell.
Currently, I have only tried two approaches to compare the reference transcripts and the SARS Cov2 sequences. Both approaches use autoencoders to get a more general comparison.
The first one relies on autoencoders and distance computations to select similar transcripts. Mean SARS Cov2 composition offers a fixed point to compare with the reference transcripts. The first round of selection is done by selecting transcripts with a distance lower than a threshold. Then the selected transcripts are compared with different samples of a latent walk. This ensures selecting transcripts that contain similar fragments as the SARS Cov2.
This approach results in the selection of around 507 transcripts. Frome those around 54 have experimental evidence to have a role with COVID-19. At this point, the screening starts to become more difficult. The scarcity of information about some of the transcripts is the primary driver. Scrapping information from Uniprot results in 135 records out of 507 lowering the odds to cluster the information by similarity.
But a subset of retrieved transcripts showed some agreement with solar radiation. An environmental variable that is tightly correlated with Covid-19 waves. Particularly some selected transcripts had a relation with vitamin D, which is synthesized by solar radiation. Low levels of vitamin D have been correlated with the complication of COVID-19. LCOR, similar to SARS Cov2, associates with HDAC6 which in turn have some regulatory role for the vitamin D receptor or VDR. SLC2536A, similar to SARS Cov2, increases its mitochondrial expression after vitamin treatment. CDS1, similar to SARS Cov2, is regulated by VDR. SLC16A10, similar to SARS Cov2, is being proposed as a response element of VDR.
The second approach tries to automate things a little bit further. The transcripts frequencies are into an autoencoder and selected by reconstruction. Sequence selection is done by low reconstruction error and z-score. This will exclude outliers and select sequences correctly reconstructed. And its reconstruction lies over the domain of the learned representation.
Using this approach with an autoencoder trained with the full SARS Cov2 sequences results in 358 sequences selected from the reference transcripts. With most of the sequences being located at the chromosomes 6, Y, and 4.

Yet, due to its size, some fragments inside the SARS Cov2 sequence might be neglected. To try to get a better segmentation sequences are split into two fragments. One that contains the non-structural genes, and another with the structural genes. This allows finding fragment-specific similarities if there are any.
The non-structural segment showed an almost continuous set of transcripts similar to the SARS Cov2 sequence. The selected transcripts showed similarity to SARS Cov2 sequences isolated at different points in time. Most of the selected transcripts were located at chromosomes 3,5 and 2.
While the structural segment resulted in the selection of transcripts that clustered together at a specific part of the learned representation. And most of the selected transcripts were located at chromosomes 4,2 and 3.

On the second approach, there was not a manual review of the selected transcripts as the election increased around 10 fold. Making it harder to find information on the selected transcripts. Yet the segmentation of the SARS Cov2 sequence appears to catch more information about the similarity between the transcripts and the viral sequences. And as the comparison is made using the autoencoder the result is a global or population comparison.
Some consequences
If susceptibility is driven by genetic mimicry and one of the first actions of the infected cell is to starve the virus by restraining the available nucleotides. Then a long-term solution could be to lower or shut down the expression of a series of genes with similar compositions. This shutdown could lead to the different symptoms experienced during the illness. Or could drive the different symptoms experienced after the acute illness.
If that is the case, then different degrees of similarity to different genes could lead to a series of apparently unrelated symptoms. As the virus adapts to the new cellular resources, then it will change how similar it is to a new subset of genes. This change will result in different symptoms or different post-acute illness sequelae.
This proposed mechanism alone is unable to explain the complete set of changes that happens during the SARS Cov2 infection, both acute and post-acute phase. However, within the small subset of changes that tries to explain it sounds like a possible explanation. Yet continuous research is needed to establish the accuracy of the presented hypothesis.